|
Contributed by Dr. Julia Kzhyshkowska
Typical modification sites: RGG box or RXR sequence motifs R-arginine, G-glycine,X-any aminoacid. Enzymes catalysing protein arginine methylation: PRMT1 from rat and the human homologue HRMT1L2, human PRMT2 (HRMT1L1), rat and human PRMT3, mouse CARM1 (PRMT4), human JBP1 (PRMT5). Substrates of PRMTs: In mammalian cells hnRNPs contain about 65% of the total NG,NG-dimethylarginine found in the cell nucleus. Additionally, functionally diverse proteins are substrates for PRMT family members, including fibrillarin and nucleolin, involved in ribosomal biogenesis, histones, fibroblast growth factor, interleukin enhancer-binding factor 3 the SRC kinase adapter protein Sam68, STAT1, and EWS.
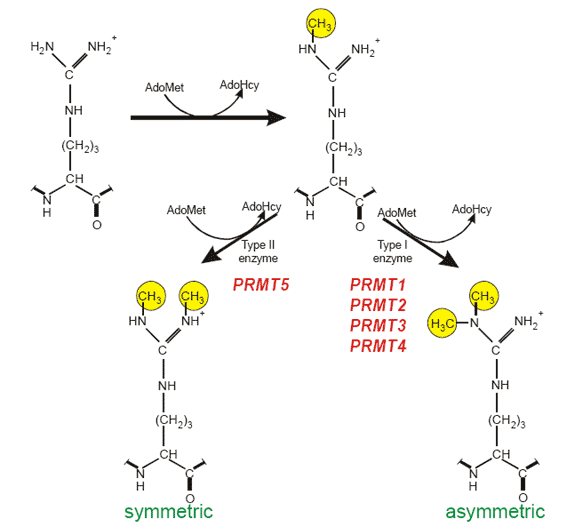
Analysis of protein arginine methylation in vivo.
- Propagate cells of interest in normal medium corresponding to the cell type (for typical adherent cells DMEM, 10% FCS, antibiotics). For adherent cells cell density on the experiment day should be 50-70%.
- Wash cells 2x with PBS and replace the medium with medium deficient in methionine (or methionine and cysteine). For standard adherent cells - MEM Eagle medium without methionine (GIBCO BRL), supplemented with penicillin, streptomycin and 5% dialysed foetal calf serum for 30 min.
- Inhibit translation by adding to the medium a cycloheximide/chloramphenicol mixture in a final concentration of 100 μg/ml and 40 μg/ml (no-translation mix, NTM), respectively.
- Incubated cells with translation inhibitors for 1h before labelling as well as during the labelling procedure. To detect methylated proteins add the methyl-group donor L-[mehtyl-3H]methionine (Amersham) in a final of concentration 30 μCi/ml.
- Monitor the protein translation inhibition by cell labelling with 35S-methionine (ICN) 20 μCi/ml in parallel control plates +/- NTM.
- Continue metabolic cell labelling for 3.5 h.
- Wash cells 2x with PBS, harvest and lyse in buffer you like. For example middle stringency buffer sufficient for lyses of adherent cells: NP40-1% lysis buffer (50 mM TRIS-HCl pH 8.0, 150 mM NaCl, 1% NP-40) supplemented with a protease inhibitor cocktail tablet (Roche).
- Identify the protein of interest by immunoprecipitation (IP) with specific antibody.
Suggested IP protocol.
- Preclear cell lysates with protein G-sepharose.
- Incubate 200μg of pre-cleared lysates overnight with protein G coupled to the corresponding antibody (rotation overnight, 4°C, total IP volume 1.5 ml)
- Wash complexes 5 times with corresponding (for example NP40-1% buffer). Keep samples at 4°C during washing steps
- Add of SDS-loading buffer were added to each sample (optimal 40-60 μl). Heat samples at 95°C for 5 min and analyse by SDS-PAGE. Incubated for 1 h with H3 enhancer, for example Enlight solution (EnerGene), dry and expose to Kodak XOMAT (AR) film at –80°C.
Do not forget important controls!
Cell lysed should be analysed in PAGE in parallel, and no signal should be observed in samples treated by NTM and labelled by 35S-methionine For more details about procedure and controls see: Kzhyshkowska J et al, Heterogeneous nuclear ribonucleoprotein E1B-AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem J. 2001 Sep 1;358(Pt 2): 305-14.

|

